CURRENT PROJECTS
Currently I am working in the lab of Dr. Daniel Bolnick studying evolutionary and ecological immunology in a fish-parasite system. The Gasterosteus aculeatus (three-spined stickleback)- Schistocephalus solidus host-parasite system is a unique example of independent repeated evolution of host immunity to a novel parasite. Approximately 12,000 years ago, sticklebacks colonized freshwater lakes where they were exposed to a cestode parasite. Populations have thus independently evolved immunity and now vary significantly in parasite resistance. Thus the stickleback-cestode system provides an excellent evolutionary immunology system which I will use to explore a number of key ecoimmunological questions.
PAST PROJECTS
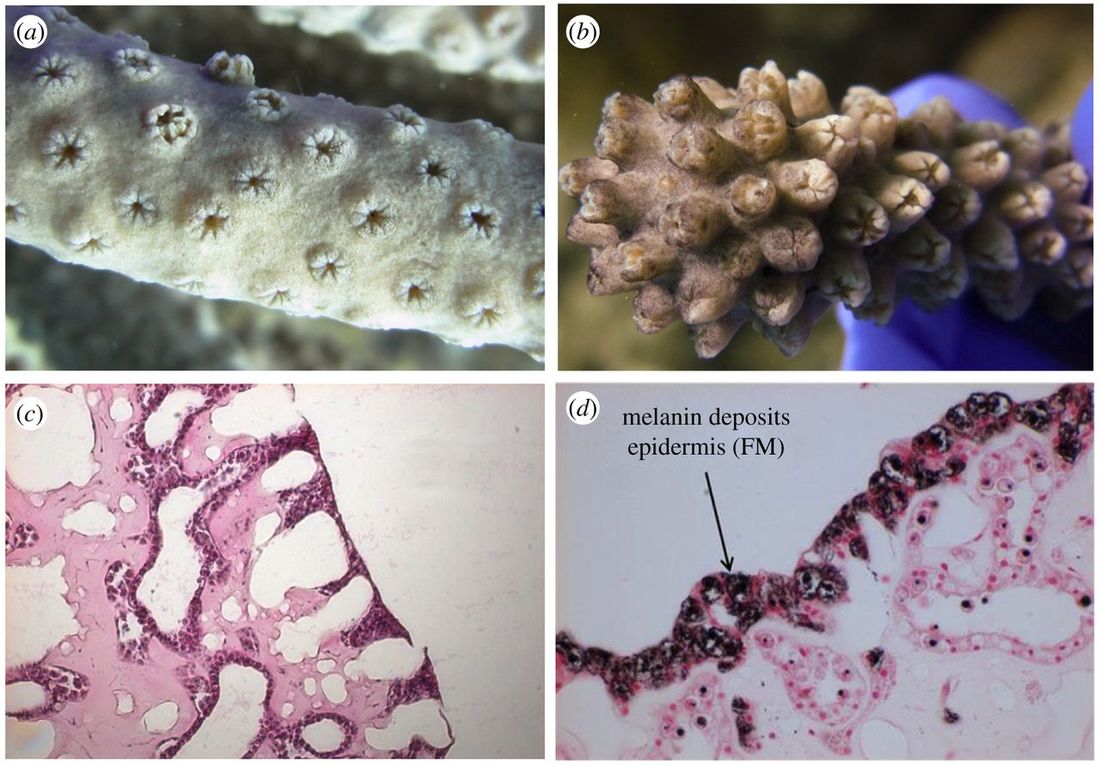
My dissertation research focused on understanding the mechanisms of and factors contributing to disease resistance in corals as well as the ecological consequences of immune response. I have studied these questions in a variety of laboratory and natural settings as well as using a variety of coral species. Predominately my research relied on transcriptional and biochemical techniques to get at differences in gene expression and protein activity which contribute to differences in immune response between coral individuals and species. Below is a list of my dissertation projects. Funding for these projects has come predominantly from NSF and from the UTA Phi Sigma Society