|
Lauren and other members of the Mydlarz and Walsh labs had another great year running an outreach booth at Texas' annual earth day festival, EarthX. They also participated in a related ocean conservation conference.
0 Comments
From October 20th to October 25th, I got the pleasure of joining a team of researchers from Rice University, Texas A&M University, and University of Houston Clearlake on a research cruise to the Flower Garden Banks (FGB). The cruise was a product of a recently funded NSF Rapid Grant to track the effects of floodwaters associated with Hurricane Harvey on the reefs at FGB. Our team included oceanographers, biologists, as well as support divers from NOAA and Moody Gardens. Researchers from Boston University will also assist in sample processing.
We spent five days upon the LUMCON R/V Point Sur conducting sampling dives for corals and sponges and collecting water quality data using CTD (Conductivity, Temperature, Depth) techniques. At times the weather didn't seem like it would cooperate, but in the end we completed all planned CTD surveys and came home with many coral and sponge samples from both banks of the perserve. The crew of the Point Sur made the ship feel like home. All in all it was a great, productive experience! I am looking forward to our portion of the project (proteomics and biochemical assays of corals) and integrating our results with all of the other groups who collected samples! As this news/blog outlet has been somewhat neglected over the past year or so, I've decided to lump my summer updates into one big post. So here are the highlights from summer field work!
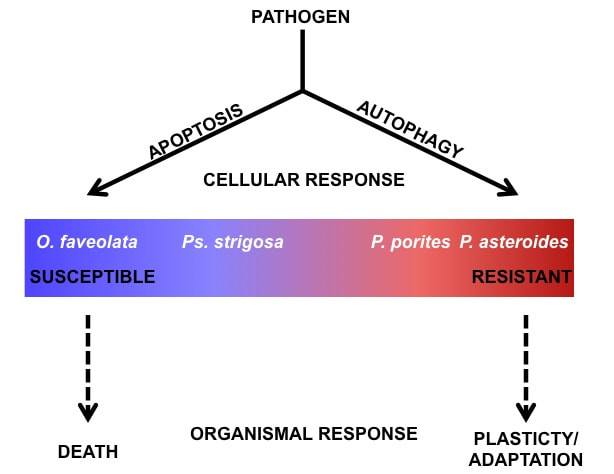
Mydlarz Lab in Roatan Honduras The Mydlarz lab (Dr. Mydlarz, Contessa Ricci, and myself) traveled to Roatan Honduras where we spent a week at the Roatan Institute of Marine Science assisting with LSAMP research conducted by a group of undergraduate students from Midland College. Lots of diving and cool research! Field Research in St. Thomas In the end of the June, the entire Mydlarz lab, including new graduate students Nick and Brad, and my undergraduate student Caleb, traveled to St. Thomas to conduct research for our newest EAGER grant. There we worked with Marilyn Brandt at the University of the Virgin Islands on a variety of projects, and I completed the field research for the last chapter of my disseration.  On September 19th, 2017, Lauren was invited to kick off the St. Edward's biology departmental seminar series. There she spoke about her ongoing research investigating coral immune response and the interplay between coral symbiosis and immunity. Over 120 undergraduate students and faculty attended the talk!! Thanks to Matt Steffenson and Dan Gold for the invitation and their amazing hospitality. It's been a while, but I wanted to take some time to highlight my newest publication in Proceedings of the Royal Society B (Life or death: disease-tolerant coral species activate autophagy following immune challenge). You can click here or go to the publication section of this website to read the newest paper. We're really excited about these findings as we believe they are particularly novel to the field of coral immunology and ecology. In short we highlight the importance of apoptosis and autophagy in coral immune response and disease susceptibility. Check it out!
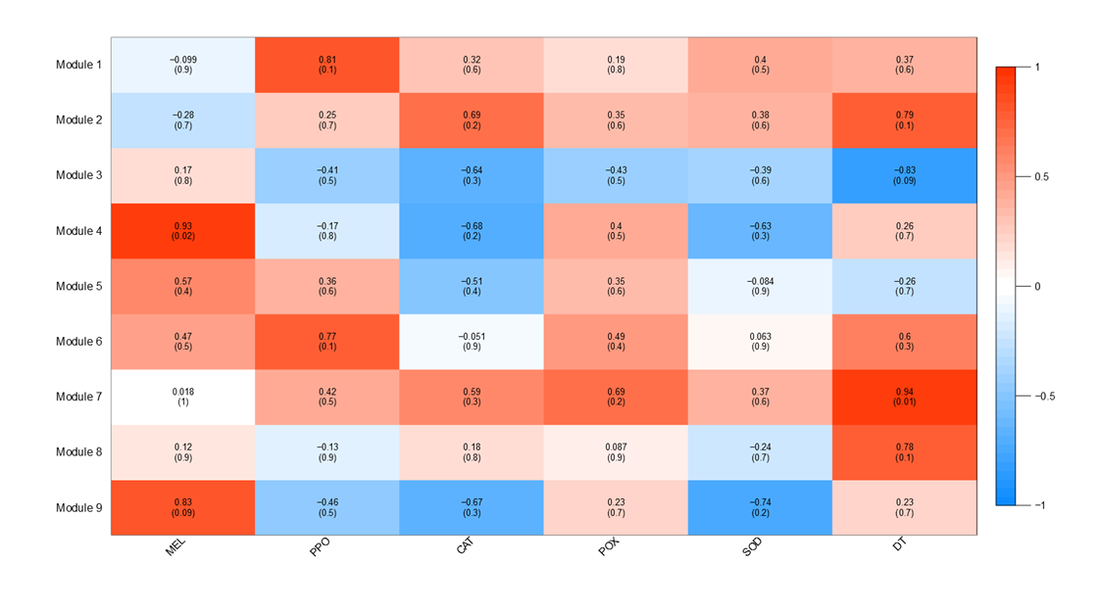
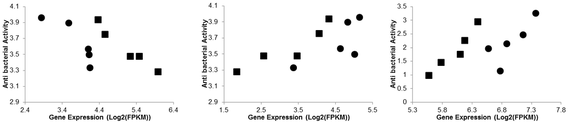
Just a few weeks ago, my newest publication was accepted for publication in Developmental and Comparative Immunology. If you haven't read it yet, you can check it out here or go to my publication page and download the pdf! To summarize the paper: we used novel correlative and network based analyses to connect gene expression to protein data. It may seem straight forward enough, but working on such things in a non-model organism was certainly a challenge! It took a lot of creativity, but in the end we were able to achieve our goal of identifying new transcripts which change in expression during immune challenge and were correlated to immune protein activity! Now that the paper's out, I thought I'd devote a blog post to detailing the journey we took to get to that final goal. When I first started my graduate career, my main goal was to go beyond the, at that time, standard in transcriptional studies of non-model organisms. I was hoping to move beyond lists of deferentially expressed genes. Namely I wanted to employ more "genome to phenome" type techniques, so to speak, in order to use transcriptomics to really get at ecological questions. This new paper was my first attempt at doing just that. At SICB in 2014, I first learned of a new R package, Weighted Gene Correlation Network Analysis, which not only groups genes based on patterns of expression, but also allows for the correlation of those groups of genes to quantitative traits. This was just the kind of tool I was looking for, so naturally I wanted to try it out. Our post-doc at the time had the same idea and was kind enough to collaborate with me on this project using a great set of data from an immune challenge experiment that had previously been conducted. In the process of analyzing this dataset using WGCNA, I learned a lot about statistics, and the short comings of working in a non-model organism for which there are few gene annotations. Essentially, while WGCNA worked great at grouping genes and correlating them to our protein assay results, the lack of gene annotations made it almost impossible to draw any significant conclusions using that software alone. But, as you can see from my supplementary tombstone plots (Figure 1), there were lots of groups of genes which were significantly correlated to types of immunity! The problem was we just couldn't characterize these groups well. Figure 1: Supplementary figure displaying correlations between modules and different measures of immunity. When we realized that WGCNA just couldn't quite get us to where we wanted to go, I once again had to get creative with my analysis techniques. This time I tried something I hadn't seen done much before: directly correlating gene expression to immune activity. WGCNA became a filter: all the genes in significant modules were screened for individual correlations to a measure of immune activity. This single gene-protein correlation technique allowed us to identify genes which not only changed during immune challenge, but also had correlations to functional measures of immunity (Figure 2). Using this technique we were able to add confidence to the identification of potential candidate immune genes beyond analysis of blast annotation results. In addition, using this combination of differential expression and correlative techniques, we were able identify genes which were down-regulated post immune challenge but were annotated as positive regulators of immunity and correlated positively to functional measures of immunity. While we still don't fully understand these patterns, its possible that disregulation of sorts of these immune genes may be contributing to differential susceptibility of corals to disease. So, while the journey may have been long, and we may have had to get creative at times, I believe this new publication really does make a strong contribution to the field of marine invertebrate immunology. Moral of the story: when using transcriptional techniques to study non-model organisms, don't be afraid to get creative and travel off the beaten path. You never know what you might find! Figure 2: correlations between immune genes (LRR domain protein, DMBT1, HSP70) and measures of immunity (antibacterial activity for the first two, propenoloxidase for the third)
It's the end of the semester, and the end of another academic year, which means I've been spending countless hours preparing year end reviews for UTA and for NSF. As tedious as this process may be, I feel like its given me some good perspective on just what I've done, and failed to do, in the past year. As a whole, I feel like graduate students, myself included, are prone to condemning ourselves for not working hard enough, not publishing enough, not 'keeping up with the Jones's' per say. And sometimes I feel like these year end reviews are the perfect medicine for that ailment. As I sit back and look over the list of achievements I've compiled to supply to my committee for evaluation, I realize what a good source of self-evaluation this process actually is. Everything of significance which I've done in the past year is now written in one place. I have one document I can use to rationally evaluate my work over the year. And while I may feel like I'm a lack-luster graduate student, I am now forced to remind myself that in the past twelve months I've published 2 papers (one is in press, but I'm counting it since I received the proofs today), presented at two conferences, and taken several opportunities to participate in broader outreach events. That, combined with the two manuscripts I'm currently writing gives me some piece of mind that I'm not a complete failure as a graduate student. But this process also allows me to critically look at where I've failed: yes I got a manuscript out, but only after two years of working on it. And several other research goals from last year have barely progressed in the past twelve months. So somehow this annual review process seems to simultaneously give me piece of mind about my progress as a graduate student, and light a fire under my butt where I've been lacking. Whether the accomplishments I've listed will be 'satisfactory' for my committee, I don't know. But I can say I'm proud of my work this past year, while at the same time ready to push myself even hard to be more productive next year. Hopefully my fellow graduate students will agree with this sentiment. Cheers to summer and another year of progress!
|
AuthorLauren Fuess is a postdoctoral researcher at the University of Connecticut working with Dr. Daniel Bolnick. Formerly she was a PhD student at the University of Texas at Arlington with Dr. Laura Mydlarz. The opinions expressed here are solely her own. Archives
September 2019
Categories |




 RSS Feed
RSS Feed
